Final Output¶
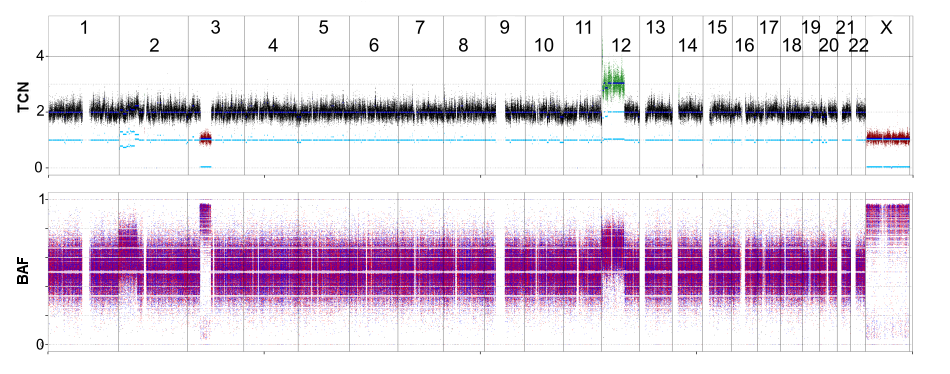
A graphical presentation of the final results is given for each tcc/ploidy solution. A general overview is give for the whole genome as shown here and per chromosome. Points represent raw SNP data points, colored by their copy number with regard to the majority copy number state (red:loss, black: neutral, green: gain). Segments are indicated by dark and light blue lines for total and allele-specific copy number respectively. Raw BAF are shown in the bottom panel which can be used to evaluate tcc and confirm allele-specific copy numbers.
Final results are provided in two formats. A reduced set of information is contained in the file ${pid}_most_important_info_${ploidy}_${purity}.txt while the full set is presented in ${pid}_comb_pro_extra_${ploidy}_${purity}.txt. A mapping of both headers and a corresponding description is given here.
| most_important_info | comb_pro_extra | description |
|---|---|---|
| chromosome | chromosome | |
| start | start | start coordinate |
| end | end | end coordinate |
| SV.Type | crest | SV type connected to both or one breakpoint |
| length | length | length of segment |
| TCN | tcnMean | total copy number |
| NbrOfHetSNPs | tcnNbrOfHets | number of control heterozygous SNPs in segment |
| dhSNPs | dhMax | estimated DH |
| minStart | minStart | last SNP prior to segment start |
| maxStart | maxStart | first SNP after segment start |
| minEnd | minStop | last SNP prior to segment end |
| maxEnd | maxStop | first SNP after segment end |
| covRatio | tcnMeanRaw | bias corrected coverage ratio |
| dhEst | dhMean | raw DH |
| c1Mean | c1Mean | minor allele copy number |
| c2Mean | c2Mean | major allele copy number |
| genotype | genotype | ratio of rounded allele copy numbers |
| CNA.type | CNA.type | DUP/DEL/LOH/TCNneutral |
| GNL | GNL | gain/loss/loh compared to diploid |
| tcnNbrOfSNPs | number of SNPs per segment | |
| tcnNbrOfLoci | number of SNPs per segment | |
| dhNbrOfLoci | heterozygous SNPs per segment | |
| map | mappable/unmappable | |
| cluster | cluster assigned during merging | |
| neighbour | indicates whether neighbouring segments exist on both sides | |
| distDH | distance to main cluster center DH | |
| errorSNP | error for DH direction | |
| distTcn | distance to main cluster center coverage ratio | |
| errorLength | error in coverage ratio direction | |
| totalError | sum of errorSNP and errorLength | |
| area | AUC ratio | |
| peaks | 1 for balanced; 2 for imbalanced | |
| meanCovT | average total coverage per cluster | |
| meanCovB | average total coverage of B allele | |
| AF | allelic factor | |
| BAF | B-allele frequency | |
| A | rounded minor allele copy number | |
| B | rounded major allele copy number | |
| TCN | rounded copy number | |
| ploidy | majority copy number used as reference for CNA.type | |
| Sex | patient sex | |
| cytoband | cytoband |